Dr Huan Liu, from Department of Periodontology, utilized machine learning to integrate functional sequencing datasets from different vertebrates revealed the conserved DNA lexicon maintaining epithelium-specific enhancer activity. This work, Analysis of zebrafish periderm enhancers facilitates identification of a regulatory variant near human KRT8/18, published on eLife on Feb 7, 2020 (https://elifesciences.org/articles/51325) could prioritize the functional study candidates from orofacial clefting genetic studies.
Orofacial clefting (OFC), including cleft lip, cleft palate, or both is among the most common structural birth defects. Such disease cause great health burden to the society. Previous studies suggested a strong genetic contribution to the etiology of OFC. Multiple genome-wide association studies (GWAS) have advanced our understanding of this contribution as multiple independent GWAS and meta-analysis have identified more than 40 associated loci. However, such statistical approaches cannot distinguish SNPs that directly influence risk from those merely in linkage disequilibrium with such SNPs. One great challenge to translate statistical association into an understanding of the biological causes of OFC is establishment of functional study pipeline.
In collaboration with Prof Robert Cornell from the University of Iowa, Dr Liu and his colleagues have been working with OFC functional study using zebrafish and mouse models. However, a mystery that has been taken for granted is why zebrafish could serve as a model for testing non-conserved non-coding variants. In his most recent studies, Dr Liu, for the first time, annotated all the active tissue-specific enhancers in zebrafish periderm, a primary epithelium, using ATAC-seq, H3K27Ac ChIP-seq, RNA-seq and transgenic zebrafish models. This work identified irf6, grhl1/3, klf17 and tfap2a/c as key transcription factors maintaining periderm differentiation and integrity. For comparison, they used the same methods to identify sets of mouse palate epithelium enhancer candidates and human oral epithelium enhancer candidates, which share a similar sets of key transcription factors. Integration using machine learning approach revealed a classifier which could prioritize OFC-associated SNPs near KRT8/18 locus. Finally, the risk alleles of top candidate SNP identified by such classifier was confirmed using reporter assay and CRISPR-Cas9 experiments.
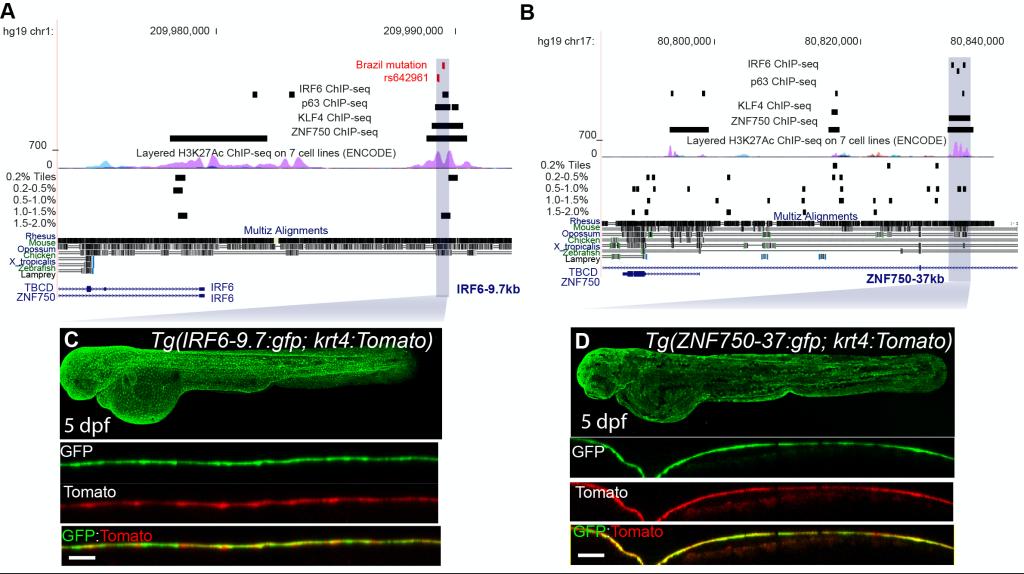
Figure A machine learning classifier trained on zebrafish periderm active enhancers applied to the human genome. A Genome browser view focused on IRF6-9.7, also known as multispecies conserved sequence MCS9.7 (hg19 chr1:209989050-209989824). B Genome browser view focused on ZNF750-37. C GFP expression pattern of Tg(IRF6-9.7:gfp; krt4:Tomato) at 5 dpf. D GFP expression pattern of Tg(ZNF750-37:gfp; krt4:Tomato) at 5 dpf.
This work was supported by grants from Natural Science Foundation of China, the Young Elite Scientist Sponsorship Program by CAST to Huan Liu and findings from NIH to Robert Cornell and Axel Visel.



